Code
# Uncomment and run this line once
#install.packages("tidyverse")
# load the package
library(tidyverse)A beginners guide to making figures in R
Noah Weidig
February 13, 2025
Nothing is worse than reading a scientific publication only to encounter muddled, overly busy figures. Clear storytelling is a cornerstone of scientific writing, and the saying “a picture is worth a thousand words” has never been more relevant. Let’s dive into creating clean, effective figures in R!
First, we need to make sure that tidyverse is installed. ggplot is a package within tidyverse, which is one of the most useful tools in R for data wrangling and visualization.
Next, lets read in our data. We’ll be working with the palmer penguins dataset, which is free and publicly available.
First, let’s look at the data in a tabular form.
# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
7 Adelie Torgersen 38.9 17.8 181 3625
8 Adelie Torgersen 39.2 19.6 195 4675
9 Adelie Torgersen 34.1 18.1 193 3475
10 Adelie Torgersen 42 20.2 190 4250
# ℹ 334 more rows
# ℹ 2 more variables: sex <fct>, year <int>So there are eight columns and 344 rows. Let’s say we are interested in penguin weight. We’ll need to look at the “body_mass_g” variable.
Let’s create a basic histogram to look at the distribution of this continuous variable.
To start, you call ggplot(). The first argument is our data.
I could also code it with data = data, although the first argument with ggplot is always the “data =” argument.
Next we have to specify the aesthetics. Since a histogram only has an x argument (our continuous variable: body_mass_g, in our case), that’s all we need.
However, this is just a blank graph. We need to specify which type of graph. In this example, we will create a histogram. Each additional “geom” is added by a “+” sign. I generally like to start a new line after each “+”.
Not bad! Now let’s customize it. I personally do not like the default ggplot look, so let’s try some built in themes.
I prefer theme_bw() or theme_minimal(). Let’s stick with those going forward.
Maybe you want to change the fill color of the histogram bars. Colors in R are always specified within quotes.
Maybe you want you want each species to have a different color. We’ll need to specify which variable is our fill within the aes().
Great! But these are stacked bars. We want them to overlap.
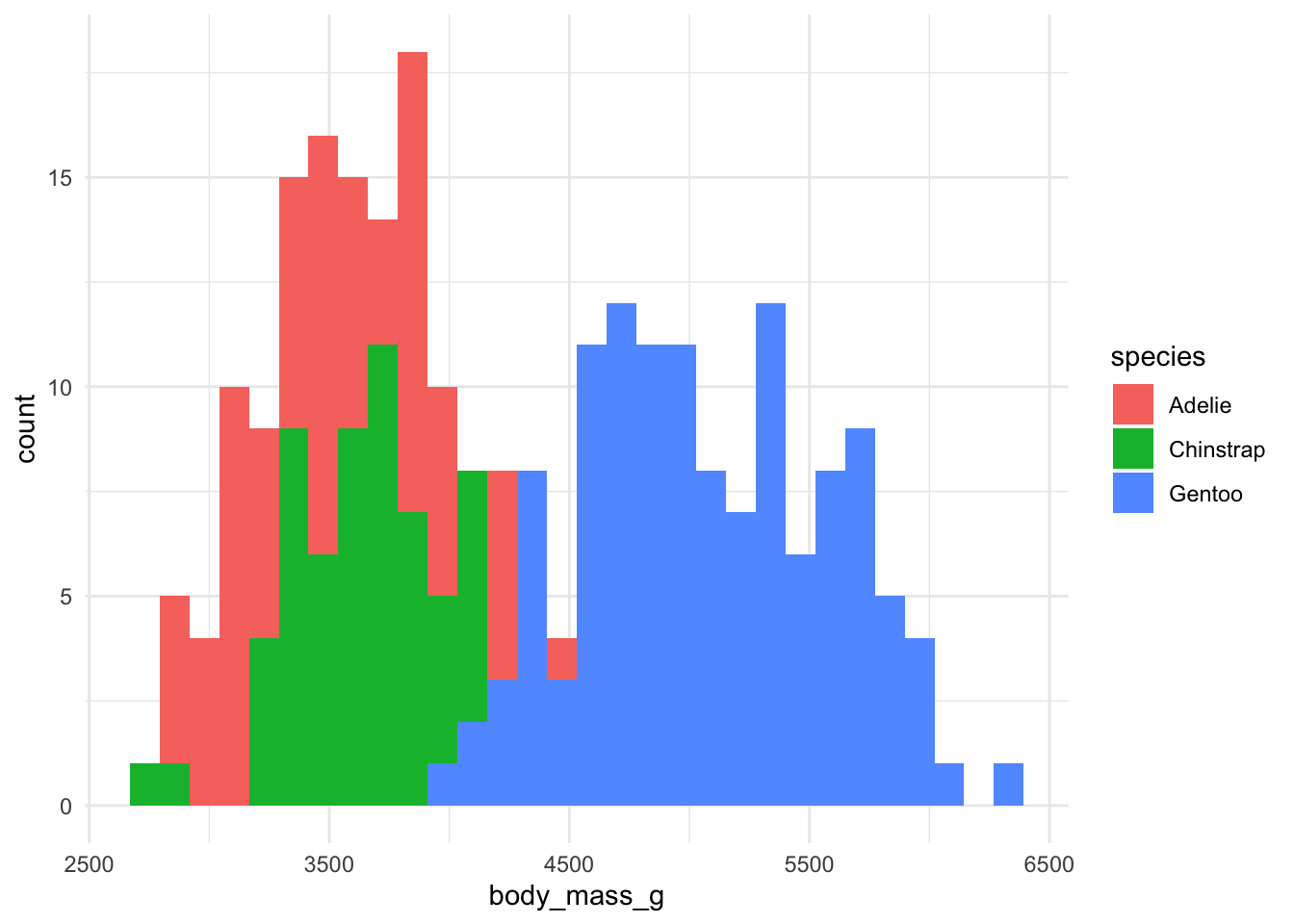
But we can’t quite see some of the bars that overlap. Let’s change the transparency (alpha).
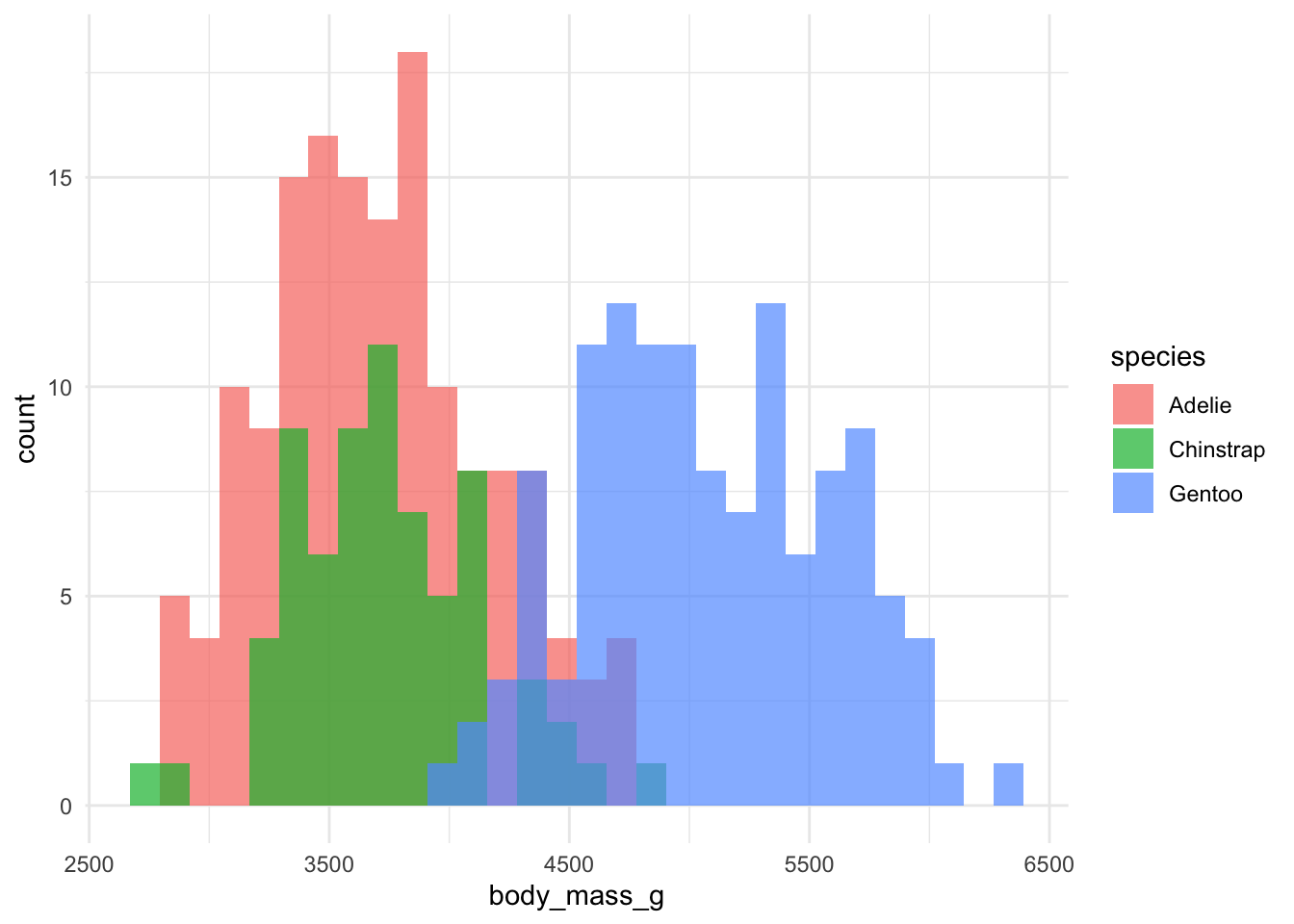
Much better!
The x and y-axes aren’t very pretty right now. Let’s change that. We’ll use labs().
Since this is a basic introduction to ggplot, we will not go into all the other geoms. However, there are several other types of graphs you can make:
geom_bar: bar graphs
geom_point: scatterplots
geom_line: linegraphs
geom_text: adding text to your figure
And so many more!
Now let’s save our figure. Most journals require between 300-600 dots per inch (dpi).
And that’s it! Thanks for reading! Look out for more R tutorials.
---
title: "Basics of ggplot2"
subtitle: "A beginners guide to making figures in R"
execute:
warning: false
author: "Noah Weidig"
date: "2025-02-13"
categories: [code, ggplot2, dataviz]
image: "image.jpg"
description: "Learn the fundamentals of ggplot2 — from basic histograms to colorful, labeled figures using the Palmer Penguins dataset."
toc: true
toc-depth: 2
code-fold: show
---
[](https://twitter.com/allison_horst)
Nothing is worse than reading a scientific publication only to encounter muddled, overly busy figures. Clear storytelling is a cornerstone of scientific writing, and the saying "a picture is worth a thousand words" has never been more relevant. Let's dive into creating clean, effective figures in R!
# Import Data
First, we need to make sure that *tidyverse* is installed. *ggplot* is a package within *tidyverse*, which is one of the most useful tools in R for data wrangling and visualization.
```{r}
# Uncomment and run this line once
#install.packages("tidyverse")
# load the package
library(tidyverse)
```
Next, lets read in our data. We'll be working with the palmer penguins dataset, which is free and publicly available.
```{r}
# Uncomment and run this line once
#install.packages("palmerpenguins")
library(palmerpenguins)
data <- penguins
```
First, let's look at the data in a tabular form.
```{r}
print(data)
```
So there are eight columns and 344 rows. Let's say we are interested in penguin weight. We'll need to look at the "body_mass_g" variable.
# Basic Syntax
Let's create a basic histogram to look at the distribution of this continuous variable.
To start, you call ggplot(). The first argument is our data.
I could also code it with data = data, although the first argument with ggplot is always the "data = " argument.
Next we have to specify the aesthetics. Since a histogram only has an x argument (our continuous variable: body_mass_g, in our case), that's all we need.
```{r}
ggplot(data = data, aes(x = body_mass_g))
# or
ggplot(data, aes(body_mass_g))
```
However, this is just a blank graph. We need to specify which type of graph. In this example, we will create a histogram. Each additional "geom" is added by a "+" sign. I generally like to start a new line after each "+".
```{r}
ggplot(data, aes(x = body_mass_g)) +
geom_histogram()
```
Not bad! Now let's customize it. I personally do not like the default ggplot look, so let's try some built in themes.
```{r}
ggplot(data, aes(x = body_mass_g)) +
geom_histogram() +
theme_bw()
```
```{r}
ggplot(data, aes(x = body_mass_g)) +
geom_histogram() +
theme_minimal()
```
```{r}
ggplot(data, aes(x = body_mass_g)) +
geom_histogram() +
theme_classic()
```
```{r}
ggplot(data, aes(x = body_mass_g)) +
geom_histogram() +
theme_linedraw()
```
I prefer theme_bw() or theme_minimal(). Let's stick with those going forward.
# Get Colorful
Maybe you want to change the fill color of the histogram bars. Colors in R are always specified within quotes.
```{r}
ggplot(data, aes(x = body_mass_g)) +
geom_histogram(fill = "red") +
theme_minimal()
```
Maybe you want you want each species to have a different color. We'll need to specify which variable is our fill *within* the aes().
```{r}
ggplot(data, aes(x = body_mass_g, fill = species)) +
geom_histogram() +
theme_minimal()
```
Great! But these are stacked bars. We want them to overlap.
```{r}
ggplot(data, aes(x = body_mass_g, fill = species)) +
geom_histogram(position = "identity") +
theme_minimal()
```
But we can't quite see some of the bars that overlap. Let's change the transparency (alpha).
```{r}
ggplot(data, aes(x = body_mass_g, fill = species)) +
geom_histogram(position = "identity", alpha = 0.7) +
theme_minimal()
```
Much better!
# Labeling Your Graph
The x and y-axes aren't very pretty right now. Let's change that. We'll use labs().
```{r}
ggplot(data, aes(x = body_mass_g, fill = species)) +
geom_histogram(position = "identity",
alpha = 0.7) +
theme_minimal() +
labs(x = "Body Mass (g)",
y = "Frequency",
fill = "Species")
```
# Other geoms
Since this is a basic introduction to ggplot, we will not go into all the other geoms. However, there are several other types of graphs you can make:
- `geom_bar`: *bar graphs*
- `geom_point`: *scatterplots*
- `geom_line`: *linegraphs*
- `geom_text`: *adding text to your figure*
- And so many more!
## Exporting Figures
Now let's save our figure. Most journals require between 300-600 dots per inch (dpi).
```{r}
#| eval: false
ggsave("penguins_histogram.jpg", width = 5, height = 3, dpi = 300)
```
And that's it! Thanks for reading! Look out for more R tutorials.